The world’s leading cause of death and disability according to World Health Organization is respiratory related infections. It is estimated that more than 1 billion people are facing the burden of respiratory related diseases.1 Diagnostic tests play an indispensable role for clinicians in drawing up a definitive diagnosis.2 The detection of varied pathogen provides information that is an essential component in conducting a prompt, appropriate and precise approach on the implementation of medical management and varied preemptive and disease control undertakings.3 According to Kha et al,4 testing and screening is vital in every clinical process, and performing improper clinical testing leads to uneconomical use of resources and places patients in peril by missing a clinical diagnosis. Hence, initial treatment is not provided, this leads to persistence of the illness, prescription of unnecessary and inappropriate antibacterial treatment leading to unwanted side effects. A decade ago, PCR was just a promising test that hoped to cater for the demand of the medical field in terms of rapid detection of potential pathogens.5 Providing appropriate medical treatment requires a definitive diagnosis thereby relying on an accurate diagnostic test. The introduction and utilization of multiplex real-time polymerase chain reaction (RT-PCR), a molecular method in detecting viral and bacterial pathogens, sheds light on the effort of having a diagnostic test that could rapidly identify the etiologic agent; thereby, leads to a timely and definitive diagnosis. The RP 33 test, a RT-PCR, performed well in a core test (global sensitivity of 75.9% and 96.5% specificity) on a study done with four varied commercial multiplex molecular tests. RP 33 showed excellent specificity values, the highest among the four panels being tested, thus proving the reliability and efficiency of RP 33 test.6 A study done in Egypt by El Baroudy NR7 posited the significance of the utilization of multiplex RT-PCR as a vital tool in reaching an accurate clinical judgement.
This is a retrospective study of the RP 33 test which took place from June 2018 to June 2019 (1 year) and was performed on twenty symptomatic participants. There were 10 males and 10 females, all were 18 years of age and older, had their checkup at OPD Chest Center in Bangkok Hospital, and their laboratory tests conducted within the duration of the study. The average age of the participants is 64 ± 8.4 years. Participants were immune competent and presented with clinical manifestations of acute respiratory infections such as fever, nasal congestion, cough, runny nose, sore throat and body malaise. The procedure was outlined to the participants and specimens for RP 33 test were taken through a nasopharyngeal swab. Additional tests, namely sputum culture and sensitivity (Sputum C/S), acid fast bacilli test (AFB test), hemo culture and sensitivity (hemo C/S), bronchial washing and Quantiferon TB test (QFT TB test) were performed on seventeen participants. Participants whose RP 33 test were undetected, required additional diagnostic tests to rule out a conclusive diagnosis. Moreover, RP 33 test that detected results but manifested unlikely signs and symptoms of acute respiratory infections (ARI) were subject to a further diagnostic test for a definitive diagnosis.
RP 33 test results often take a shorter turnaround time, a mean time of 20 ± 7.767 hours compared to 72 hours from sputum culture or blood culture. Both bacterial and viral pathogens were the prevalent etiologic agent established from the twenty participants with Staphylococcus aureus andKlebsiella pneumonia as the leading bacterial etiologic agent and Rhinovirus (RV) for the viral etiologic agent.
The pathogens with bacterial etiology were Staphylococcus aureus (S. aureus) with three cases, Klebsiella aneumonia (K. aneumonia) three cases, two cases and one case of Haemophilus anfluenza (H. influenza) and Streptococcus pneumonia (S. ineumonia) respectively. While for viral etiology, there were three cases of Human Rhinovirus (RV), two cases of Influenza A virus, one case of Influenza B virus, one case of human Corona virus OC43 (HCOV-OC43), one case of human Parainfluenza virus (HPIV) and one case of human Metapneumovirusi A/B (HMPV A/B). The detected cases of RP 33 test were attained from twelve participants, including five (25%) cases that revealed multiple types of pathogens in a single RP33 test indicating co-infection. Eight cases of undetected results: seven cases were true negativeincluding the case of pulmonary Cryptococcosis, Pseudomonas aeruginosa, Enterobacter aerogenes and Mycobacterium tuberculosis.
True negative results were validated by undetected RP 33 test and a negative additional diagnostic test (sputum C/S, AFB test, hemo C/S and bronchial washing, Quantiferon TB test) while false negative has an undetected RP 33 test and a succeeding test of sputum C/S, AFB test, hemo C/S and bronchial washing that revealed an infectious agent that is included in the RP 33 list of pathogens. One case of false negative supported by the result of sputum C/S showed K. Pneumonia.
Table 1: Viral and bacterial pathogens established by Respiratory Pathogen 33 test and their number of cases including cases of co-detection
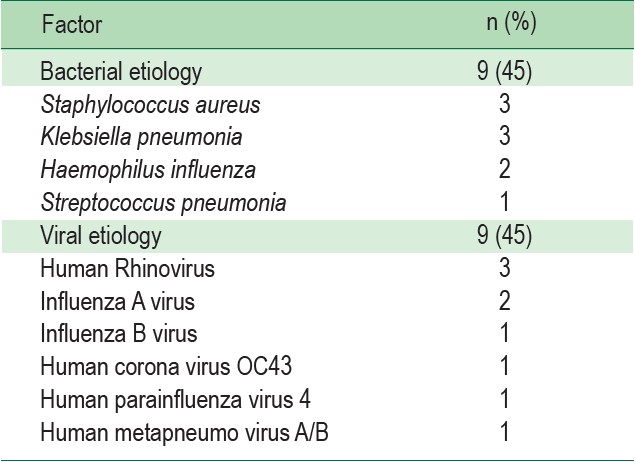
Table 2 : The list above shows the multiple types of pathogens detected in one single RP33 test

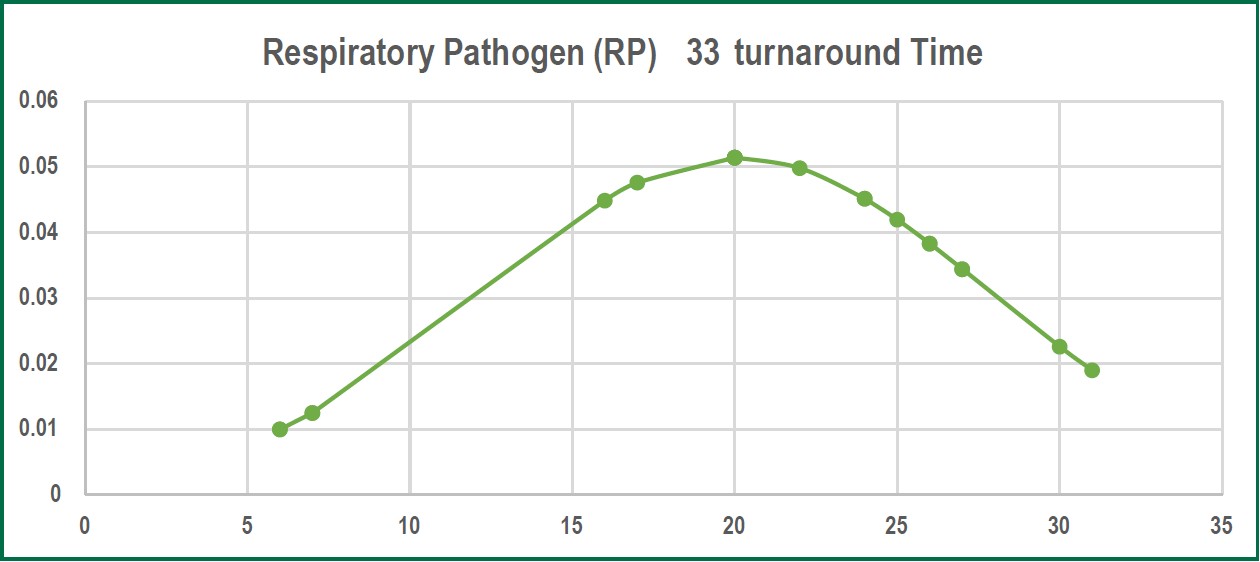
Figure 1: The graph above illustrates the normal distribution of data on turnaround time of RP33 test.
The average result on the turnaround time of RP 33, as established in our study, has a mean time of 20 ± 7.767 hours compared to 72 hours from sputum culture or blood culture. A study done by Bauer et al8 stated that at least 48-72 hours is the required time for standard technique identification of an organism, this substantiated the result of our study regarding the 20 hours mean turnaround time of RP 33 test. There are no significant differences between saliva or nasopharyngeal swab which is in concordance with the study done by Kim YG et al9 that stated none of the sampling methods (saliva or nasopharyngeal swab) provided a constant result in the context of sensitivity. Therefore to perform the RP 33 test, our studyutilized nasopharyngeal swab due to ease of accessibility. Obtaining a sputum sample takes a considerable amount of time for the patient, this leads to extended waiting time for results, and therefore, ruling out of diagnosis in a timely manner is affected, which as a consequence halts the initiation oftreatment and perhaps alters or provides erroneous laboratory results.10 The result of our RP 33 test established 90% detection of both bacterial and viral etiologies, molecular tests can detect influenza viral RNA for a longer period of time compared to other influenza tests. In our study none of thesucceeding laboratory test results (sputum C/S, AFB test, hemo C/S, bronchial washing, influenza AB&RSV and Quantiferon TB test) showed concordance with the etiologic agents detected in RP 33 test. The additional laboratory test revealed varied types of bacteria that are not within the RP 33 list of pathogens. There are several factors that might directly influence this outcome, according to Center for Disease Control and Prevention (CDC) stated that longer than 72 hours after onset of symptoms and post exposure to antibiotic regimen lessens the probability of viral and bacterial detection.11-13 The detected etiologic agents in RP 33 test demonstrated a significant association of commensal microbiota to the clinical manifestations and etiology of certain diseases, and14 our study revealed both bacterial and viral etiology. The results of the RP 33 test among twenty participants revealed S. aureus, K. pneumonia and Rhinovirus as the leading cause of ARI. S. aureus as one of the leading etiologic agents detected in our study has long been known as a normal micro-flora in the human body yet beyond its harmless state is its refutable image of having a potent capacity to cause countless cases of morbidity and mortality.15,16 Moreover, the K. pneumonia occurrences in this study on the one hand is also substantial. A study of Paczosa et al17 postulated that K. pneumonia “causes infections at a variety of sites in humans”. A keen interpretation and analysis on the degree of viral influences towards respiratory infection is essential in planning a treatment that focuses on the patients’ diagnosis; thus, avoiding unnecessary prescription of anti-microbial drugs. Rhedin et al.18 identify Rhinovirus as the most common etiologic agent that causes acute respiratory illness. The next pathogen with the greatest number is Haemophillus influenza. A study done in United Kingdom by Moxon ER et al.19 states that the etiologic agent of ARI (pneumonia) and meningitis cases are caused by Haemophillus influenza. Moreover, there were RP 33 results that revealed more than one pathogen thus indicating co-infection. A research of Cebey-Lopez et al.20 suggested there is less probable concordance between co-infection and severity of a respiratory disease but stated bacterial super infection influences the severity of disease progression. The crucial role of RP33 includes providing a rapid and conclusive identification of pathogens based on the mean result in turnaround time as indicated in our study, which reduces the needed time in ruling out diagnosis, furthermore it significantly saves the patient from unwarranted pharmacologic side effects and eases financial burden.
Respiratory Pathogen (RP) 33 test, a multiplex real time PCR, provided a commendable implication. RP 33 test provided a prompt and optimal diagnostic evaluation in ruling out etiologies oxf respiratory related infections particularly acute respiratory infections thus delivering definitive and precise clinical care to patients leading to minimal usage of broad spectrum antibiotics.