KRAS is an oncogene located on the short arm of chromosome 12 (12p12.1). The gene is a member of the Ras family of small guanine nucleotide–binding proteins, first identified as a cellular homolog of a transforming gene in the Kirsten rat sarcoma virus.1 Activating mutations in KRAS have been noted in a variety of human cancers, including carcinomas of the pancreas, lung (non-small cell lung cancer), and colorectum (Table 1). KRAS mutations are frequently found in exon 1 (codons 12 and 13) and exon 2 (codon 61).2 Mutations in KRAS codons 12 and 13 have been associated with lack of response to EGFR-targeted therapies in patients with colorectal cancer (CRC)(Table 2).3, 4
In 2009, The American Society of Clinical Oncology (ASCO) issued their first provisional clinical opinion recommending that all patients with CRC who are candidates for anti-EGFR monoclonal antibody therapy should undergo testing for KRAS mutations and that patients with KRAS should not receive anti-EGFR monoclonal antibody therapy as part of their treatment.5 Common 7 somatic mutations in KRAS codons 12 and 13 have been recommended for evaluation of KRAS gene mutation in CRC (Table 3).3-5
Table 1: Frequency of KRAS Mutations in Human Cancers1
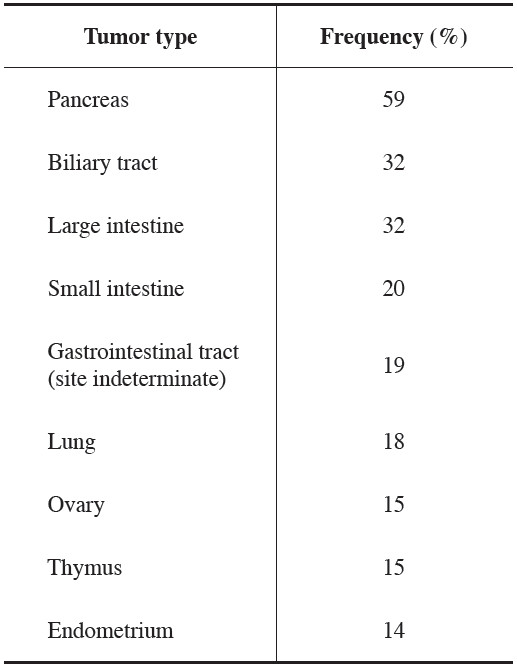
Table 2: Impact of KRAS Mutations in Patients with Metastatic CRC Treated with EGFR

Table 3: Common 7 KRAS mutations in exon 1 (codons 12 and 13) in CRC
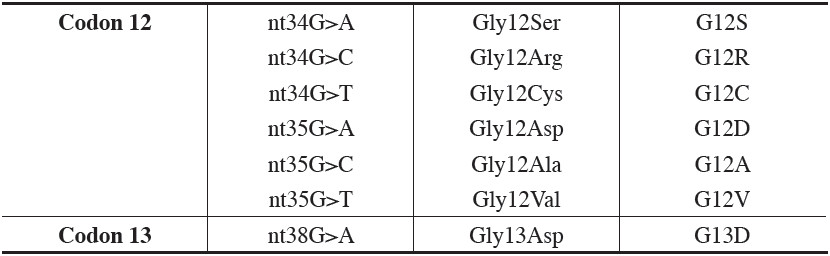
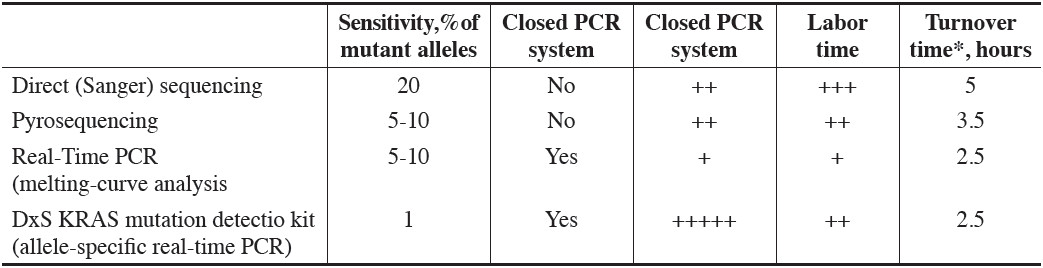
Table 4: Comparison of Methods Used for KRAS Mutation Testing 2, 6
KRAS plays a pivotal role in the EGFR signaling network. When epidermal growth factor occupies the EGFR, it activates a signaling pathway cascade through the downstream effectors of the mitogenactivated pro- tein kinases (MAPK) pathway. KRAS is one of the effectors (in addition to BRAF, ERK, and MAPK) which influence cellular proliferation, adhesion, angiogenesis, migration, and survival1,6 Blocking EGFR with mono- clonal antibodies (cetuximab or panitumumab) inhibits all downstream effects of the receptor. However, if the signaling pathways are activated independent of EGFR, as happens when the KRAS gene is mutated, these anti- EGFR agents become ineffective.1, 6 Mutation of other genes in the downstream pathway such as BRAF could result in the same effect.6
Detection of KRAS mutations in colorectal cancer tissue can be done by several molecular methods. Commonly used techniques can be divided in 2 main categories: DNA sequencing and real-time PCR (poly- merase chain reaction).
DNA Sequencing Direct (Sanger) sequencing and pyrosequencing can be used for KRAS mutation testing. While the former has been considered a gold standard, the technique is time-consuming and the sensitivity is rather low (20% of mutant alleles required for detection) (Table 4). Pyrosequencing (Figure1) is faster than Sanger, with a lesser amount of mutant alleles required for mutation detecting.6
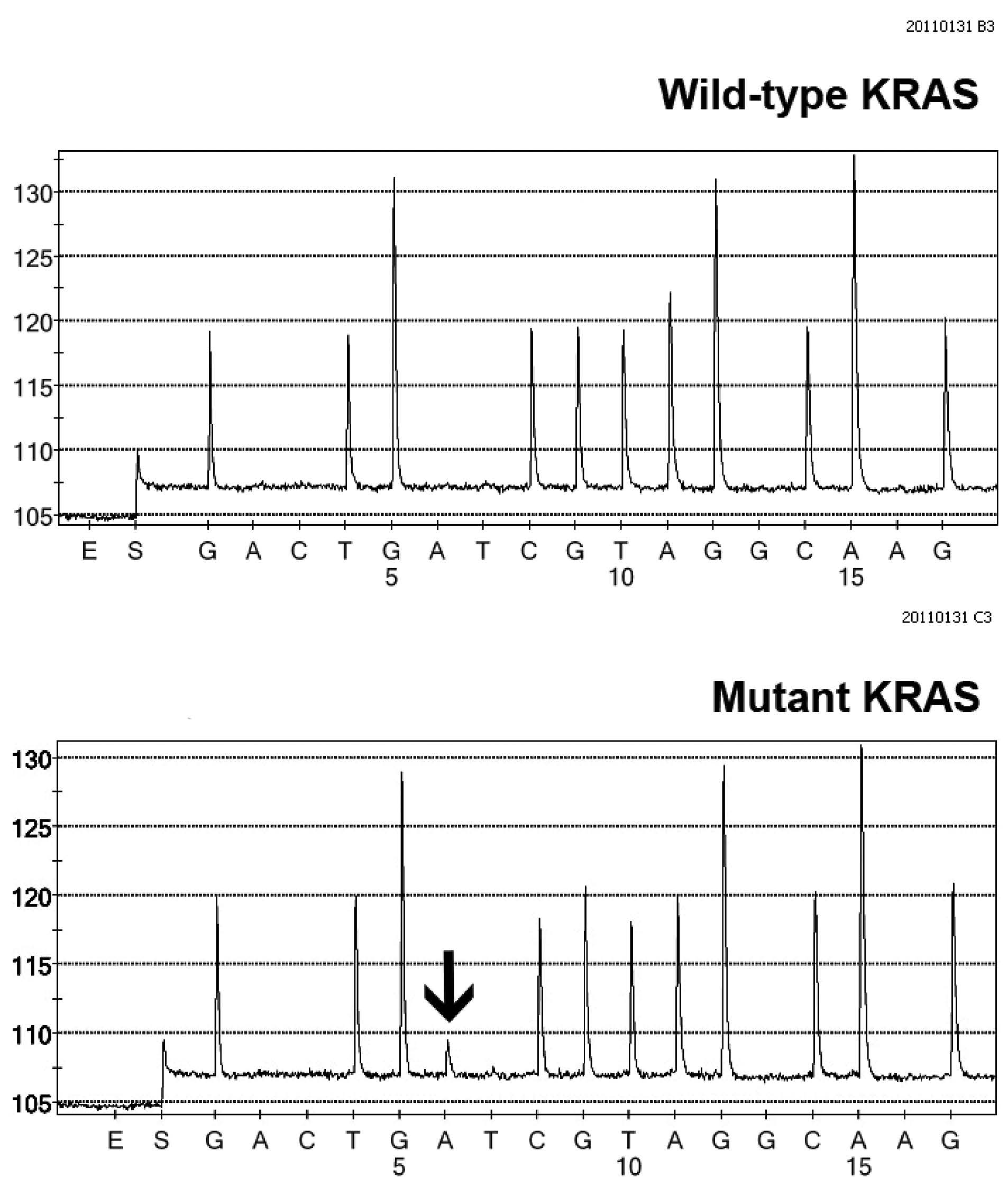
Figure 1: KRAS testing using pyrosequencing. Pyrograms demonstrate wild-type KRAS gene (upper panel) and mutant KRAS gene (lower panel). Note a mutant “A” peak (arrow) indicating nt38 G>A (Gly13Asp, G13D) at codon 13, exon 1 of the KRAS gene. (Courtesy of Dr Chupong Ittiwut, PhD, Chulalongkorn GenePRO Center, Faculty of Medicine, Chulalongkorn University)
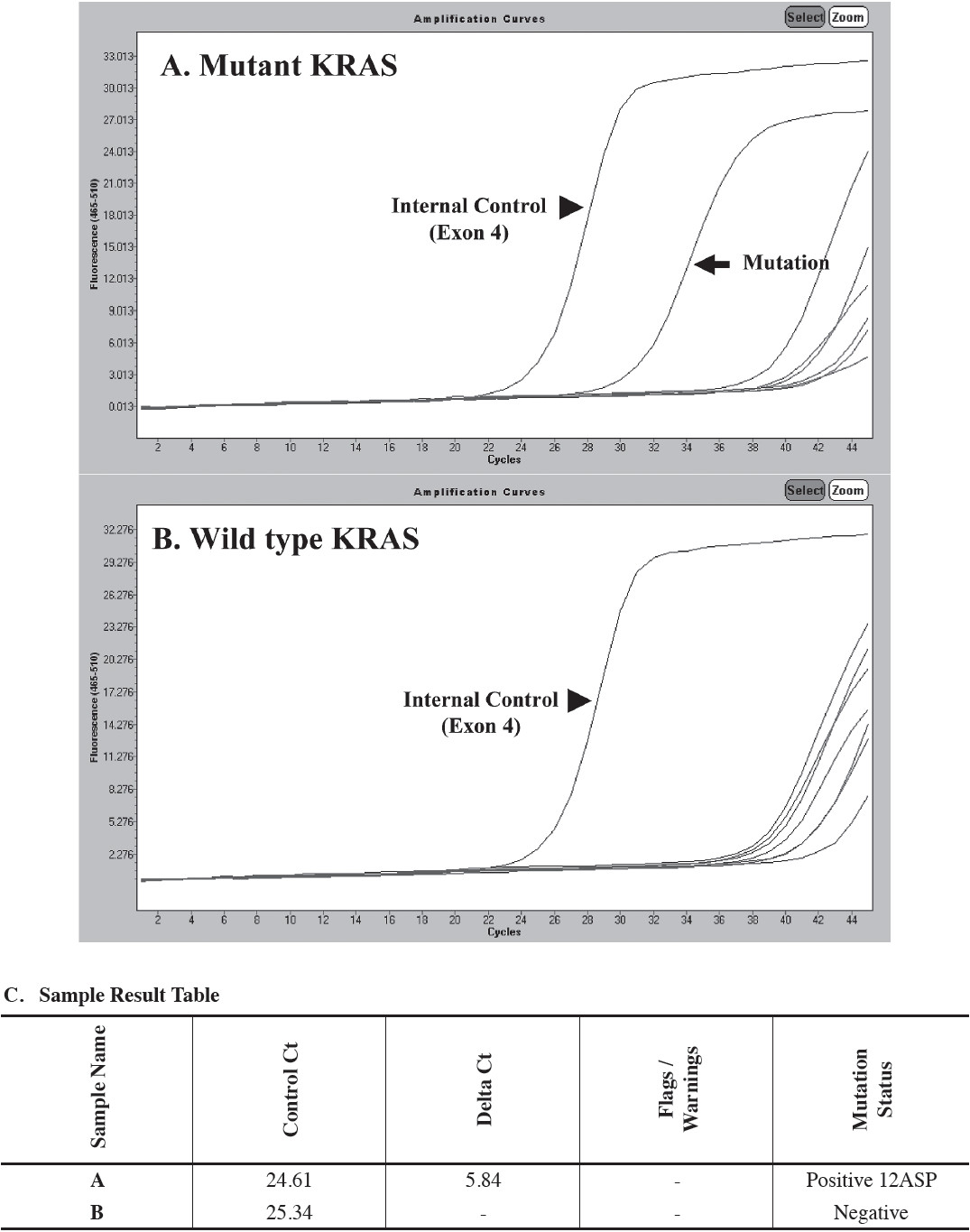

Figure 2: KRAS testing using allele-specific real-time PCR (The DxS TheraScreen KRAS Mutation Kit). (A) Positive for KRAS mutation. Note the second curve (arrow) representing the amplification of a KRAS mutation. (B) Negative for KRAS mutation. Note the curve at cycle 24 (arrowhead) served as internal control (exon 4 of the KRAS gene). (C) The DxS TheraScreen KRAS Mutation Kit analysis report using the LightCycler® Adapt Software v1.1 (Roche Diagnostics, Penzberg, Germany). (Courtesy of Dr. Alisa Junpee, PhD, Bio Molecular and Pathology Laboratory, National Healthcare Systems CO., Ltd)
Real-Time PCR The presence of a KRAS mutation can be detected either by allele-specific real-time PCR amplification (Figure 2) or by post-PCR fluorescent melting-curve analysis. Both techniques are closed PCR system, thus reducing risk of contamination. The former is available as a commercial kit, and it is estimated to be able to detect only 1% of mutant allele.6 However, the reagent cost is very high, and the technique requires more tissue for analysis as compared with other methods.2, 6 The melting-curve analysis has the similar sensitivity as does the pyrosequencing, but distinguishing among mutation types can occasionally be problematic.6
115 and 153 colon cancer specimens from Thai patients have been tested for KRAS mutations at Bio Molecular and Pathology Laboratory, National Health- care Systems CO., Ltd by allele-specific real-time PCR (TheraScreen kit, DxS Ltd, Manchester, UK) and Chulalongkorn GenePRO Center (pyrosequencing), respectively (data kindly provided by Drs Chupong Ittiwut, PhD, and Alisa Junpee, PhD). 31.3% (36/115) and 37.9% (58/153) of cases were found to carry KRAS gene mutation, total mutant KRAS cases = 37.3% (94/268). Gly12Asp (G12D), Gly12Val (G12V), and Gly13Asp (G13D) are among the commonest KRAS mutation identified in both centers. The overall percentage of KRAS mutation found in our Thai patients is within the range (30-40%) previously described in the literature.6
The most challengers in oncology is that of patient selection for therapy with molecularly targeted agents. Kras are important determinants of response or resistance to anti EFGR antibodies.