Acinetobacter baumannii, a Gram-negative coccobacilli, is a majorcausative pathogen involving nosocomial infection in various organs,such as the lower respiratory tract, skin/soft tissue, blood and, rarely,in the urinary tract and central nervous system.1 In Thailand, regarding thedata of the first half-year of 2018,2 A. baumannii was the third and secondranked organism isolated from all specimens and sputum, respectively.
This pathogen is not only an important nosocomial pathogen, but alsohas multiple mechanisms to resist the current antimicrobials varied frommulti-drug treatment (more than three groups), extensive drug resistance(resistant to all antibiotics except colistin and tigecycline) to pan-drug(resistant to all available antibiotics).3 Over the past 19 years (from 2000-2018),the prevalence of carbepenem resistant A. baumannii (CRAB) has increased.2The National Antimicrobial Resistance Surveillance, Thailand (NARST)reported that among A. baumannii isolates from hospitalized patients in 50 hospitals, the rate of CRAB increased from 5.8% in 2000 to 52.5% in 2018.2
However, the increasing rate of CRAB has affected carbapenems use asempirical therapy for infections suspected of A. baumannii. The known typeof carbapenemase enzyme in CRAB remains important for somecircumstances including the role of carbapenems in combination with theother antimicrobials against CRAB and the upcoming use of new betalactamaseinhibitor such as avibactam against carbapenemase producingorganisms.4,5
To date, carbapenem-destroying enzymes in CRAB have been found intwo major types, namely, OXA-carbapenemases and metallo-beta lactamases.1 In Thailand, several reports have revealed that blaOXA23constituted the majority of carbapenemase genes6,7 but therare prevalence of OXA-408 and IMP-19 among CRABclinical isolates was observed. However, previous studies wereconducted in university hospitals and a general hospital. Theresistant mechanisms in CRAB from a private hospital mightbe different regarding the patterns of antibiotic use, patientcharacteristics and infection prevention and control methods.Thus, our study aimed to identify the presence of carbapenemasegenes and clonal relationship among CRAB strains isolatedfrom patients admitted to a private hospital.
Bacterial strains
All clinical A. baumannii strains were obtained frompatients admitted to Phyathai II International Hospital, a 550-bedprivate hospital between August 2014 and April 2015. Acarbapenem resistant strain was defined as isolates that werephenotypically resistant to imipenem (10 μg) and meropenem(10 μg) using the disk diffusion method based on The Clinicaland Laboratory Standards Institute (CLSI).10 Only the firstCRAB isolate from each patient was kept in tryptic soy brothcontaining 20% glycerol at -80°C until studied. The researchprotocols were approved by the Ethic Committee [No.ID0014/59].
Carbapenemase genes
Each DNA sample of CRAB strains was extracted usinga commercial kit (RBC Bioscience, California, USA). Theprimers of genes (blaOXA23, blaOXA40, and blaOXA58) and conditionsthat were used, are described in Table 1. Thermocycler wasperformed as follows: 94ºC for 5 minutes; 30 cycles of 94ºCfor 45 seconds, annealing temperature specific for eachprimer pair for 45 seconds, and 72ºC for 1 minute; with a finalheating at 72ºC for 10 minutes.8
The detection of carbapenemase genes including blaIMP,blaVIM, blaKPC, blaOXA48 and blaNDM was performed, however,with multiplex PCR (Table 1). Amplification was carried outwith the following thermal cycling conditions: at 94°C for 10minutes and 36 cycles of amplification consisting of 30 secondsat 94°C, 40 seconds at 52°C, and 50 seconds at 72°C, with 5minutes at 72°C for the final extension.11
All amplicons were separated by agarose gel electrophoresis,stained with ethidium bromide, and compared with those ofknown carbapenemase genes. Finally, their identities wereconfirmed by nucleotide sequencing (Ward Medic, Ltd, Bangkok,Thailand) and were compared with known sequences in theGenBank database.
Table 1: Primers, amplicon sizes and annealing temperature used in PCR-based detection ofA. baumannii carbapenemase genes. 8,11
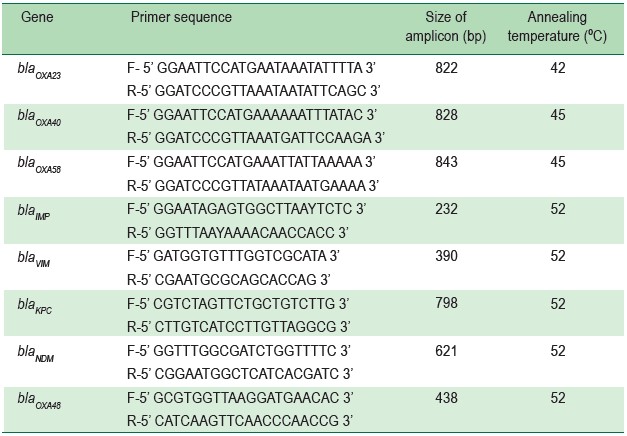
Clonal relationships
The clonal relationships of CRAB were evaluated usingthe REP-PCR method. The 15 μl PCR mixture was composedof 1 μl of DNA, 0.4 μl of 20 μM each forward and reverseprimers, 7.5 μl of PCR master mix kit (JumpStart Red Taq® Ready Mix, California, USA) and 5.7 μl of DNAase-freewater. A couple primer (REP-forward: 5’-IIIGC GCCGICATCAGGC-3’ and REP-reverse: 5’-ACGTCTTATCAGGCCTAC-3’) was used to amplify the REP region under thefollowing conditions, starting with heating at 94ºC for 10 minutes, followed by 30 cycles of 94ºC for 1 minute, 45ºC for 1 minute, 72ºC for 2 minutes and finally at 72ºC for 16 minutes 8.The REP-PCR products were performed using agarose gelelectrophoresis and were stained with ethidium bromide. Thecriterion for classifying the different clones was a pattern thatdiffered from the at least three bands or more of REP-PCR.12
Over the nine-month study period, only the first CRABstrain isolated from each patient, for a total of 15 clinicalisolates were determined. The 15 samples were isolated fromsputum (n = 9), blood (n = 2), urine (n = 1), pus (n = 2) andtissue (n = 1) specimens. All CRAB strains resisted toceftazidime and were susceptible to ciprofloxacin, amikacin,and ampicillin/sulbactam at 6.7%, 26.7%, and 26.7%,respectively.According to the clonal relationship study, the CRABisolates were categorized by REP-PCR in 8 groups [A-H], with53.3% belonging to group A, whereas the remaining 7 cloneswere in each member of B-H, respectively. (Figure 1). Of theOXA and MBL genes identified, most CRAB carried only blaOXA23 (86.7%) whereas only two isolates harbored both blaNDM1 and blaOXA23 (13.3%) (Figure 2). However, no blaOXA40,blaOXA58, blaIMP, blaVIM, and blaKPC were identified in the study.
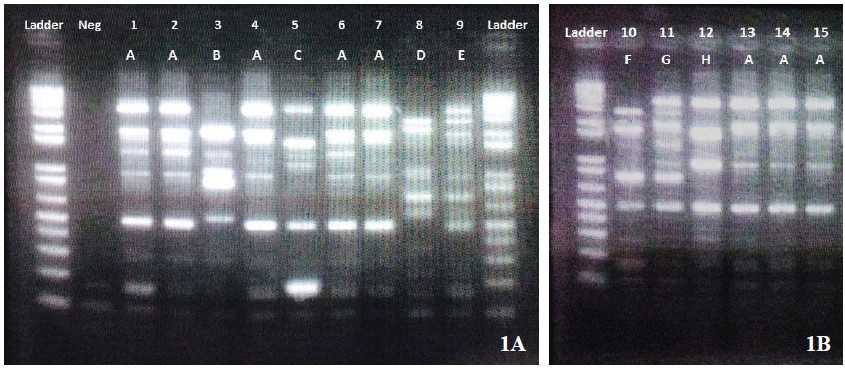
Figure 1A-B: Pattern obtained with Repetitive Extragenic Palindromic-Polymerase Chain Reaction (REP-PCR), The lettersabove each lane indicate the strain (No 1-15); Ladder, DNA molecular weight marker in kilobase unit (kb); Neg, Negative control.Using the REP-PCR method, there were 8 groups divided into patterns A-E (1A) and patterns F-H (1B).

Figure 2: PCR detection of presence of NDM-1 gene in Acinetobacter baumannii isolate No 3 and 4. Ladder, molecular sizemarkers (size (bp) is indicated in the left margin); Neg, negative control; IMP positive control; VIM positive control; OXA-48positive control; NDM positive control; KPC positive control; 1-10, test samples.
Currently, with the mechanisms of resistance in A. baumanniiespecially, carbapenemases has been reported from variousparts of the world. Although six studies detected carbapenemasegenes in Thailand only five were from the clinical isolates inthe university hospitals and the remaining were from a generalhospital. 6-9,13,14 Actually, the diversity of resistant mechanismsamong A. baumannii clones might be possible in differenthospitals.15 Moreover, at the same hospital but in differentwards, the distribution of clones and mechanism of resistancealso varied in different clinical departments.16 Thus, our study,performed in a private hospital, revealed the same blaOXA23 asrelated studies but reported two patients carrying twocarbapenemase genes (blaOXA23 and blaNDM1) simultaneously.
With the NDM-1, the Amber class B, MBL group, is oneof the most commonly reported among Enterobacteriaceae,being firstly identified in a patient who had returned from NewDelhi.17 BlaNDM-carrying Enterobacteriaceae remains on theIndian subcontinent, but to date, has been found in variousparts of the world.17 Of blaNDM-blaOXA23 carrying A. baumannii,the co-carbapenemase genes found in our study, this phenomenawas similar to a related study showing the coexistence ofblaOXA-blaNDM1 among three isolates of CRAB in India and twoisolates of CRAB in Thailand.18,19 However, we could notexplain how these genes coexisted. However, we hypothesizethat the co-genes might have been transferred by mobilegenetic elements within ISAba1.18
However, no other carbapenemase genes (blaOXA48, blaKPC,blaIMP, or blaVIM) were detected in the present study. This mightbe due to the small sample size, limited period of samplecollection, or their extremely low prevalence in the hospitalsetting. This limitation should be corrected by further studies witha larger sample size and a longer study period.
At the time of writing, avibactam and vaborbactam arediazabicyclo-octane and cyclic boronic acid respectively,having an inhibitor activity against class A, class C and someenzymes in class D beta-lactamases. Whereas class Bmetallo-beta lactamases (such as NDM, IMP, VIM) haveproven to have less inhibition by avibactam and vaborbactam.Thus, the presence of a pathogen carrying co-carbapenemasegene (blaOXA23 and blaNDM1) is challenging treatment forfinding a novel β-lactamase inhibitor with high affinity to allclasses of beta-lactamases.20
Regarding the clonal relationships in this study, clone A(53.3%) was predominant. This prevalence of the majorityclone was less than that reported in a related study (93.0%).8The lower prevalence of the predominant clone and the numeroustypes of clone might have stemmed from well-controlledmultiple factors including strict infection control guidelines,appropriate use of antibiotics and notification of infectedpatients.21 However, as clone A exhibited the highest prevalence,we suggested that the infectious control program couldcontinue minimizing the reservoir for bacterial transmissionin the hospital.22
This research comprised a study to confirm the mostcommon type of blaOXA23 found in Thailand beyond academicmedical centers. Additionally, this study firmly showed blaNDM1coexisted with blaOXA23 in clinical CRAB isolates in Thailand.
The authors declare no conflict of interest.